10.10 Plotting the data and model fits
We can plot the fits for our DFA model along with the data. The following function will return the fitted values ± (1-\(\alpha\))% confidence intervals.
get_DFA_fits <- function(MLEobj, dd = NULL, alpha = 0.05) {
## empty list for results
fits <- list()
## extra stuff for var() calcs
Ey <- MARSS:::MARSShatyt(MLEobj)
## model params
ZZ <- coef(MLEobj, type = "matrix")$Z
## number of obs ts
nn <- dim(Ey$ytT)[1]
## number of time steps
TT <- dim(Ey$ytT)[2]
## get the inverse of the rotation matrix
H_inv <- varimax(ZZ)$rotmat
## check for covars
if (!is.null(dd)) {
DD <- coef(MLEobj, type = "matrix")$D
## model expectation
fits$ex <- ZZ %*% H_inv %*% MLEobj$states + DD %*% dd
} else {
## model expectation
fits$ex <- ZZ %*% H_inv %*% MLEobj$states
}
## Var in model fits
VtT <- MARSSkfss(MLEobj)$VtT
VV <- NULL
for (tt in 1:TT) {
RZVZ <- coef(MLEobj, type = "matrix")$R - ZZ %*% VtT[,
, tt] %*% t(ZZ)
SS <- Ey$yxtT[, , tt] - Ey$ytT[, tt, drop = FALSE] %*%
t(MLEobj$states[, tt, drop = FALSE])
VV <- cbind(VV, diag(RZVZ + SS %*% t(ZZ) + ZZ %*% t(SS)))
}
SE <- sqrt(VV)
## upper & lower (1-alpha)% CI
fits$up <- qnorm(1 - alpha/2) * SE + fits$ex
fits$lo <- qnorm(alpha/2) * SE + fits$ex
return(fits)
}Here are time series of the five phytoplankton groups (points) with the mean of the DFA fits (black line) and the 95% confidence intervals (gray lines).
## get model fits & CI's
mod_fit <- get_DFA_fits(dfa_1)
## plot the fits
ylbl <- phytoplankton
par(mfrow = c(N_ts, 1), mai = c(0.5, 0.7, 0.1, 0.1), omi = c(0,
0, 0, 0))
for (i in 1:N_ts) {
up <- mod_fit$up[i, ]
mn <- mod_fit$ex[i, ]
lo <- mod_fit$lo[i, ]
plot(w_ts, mn, xlab = "", ylab = ylbl[i], xaxt = "n", type = "n",
cex.lab = 1.2, ylim = c(min(lo), max(up)))
axis(1, 12 * (0:dim(dat_1980)[2]) + 1, yr_frst + 0:dim(dat_1980)[2])
points(w_ts, dat[i, ], pch = 16, col = clr[i])
lines(w_ts, up, col = "darkgray")
lines(w_ts, mn, col = "black", lwd = 2)
lines(w_ts, lo, col = "darkgray")
}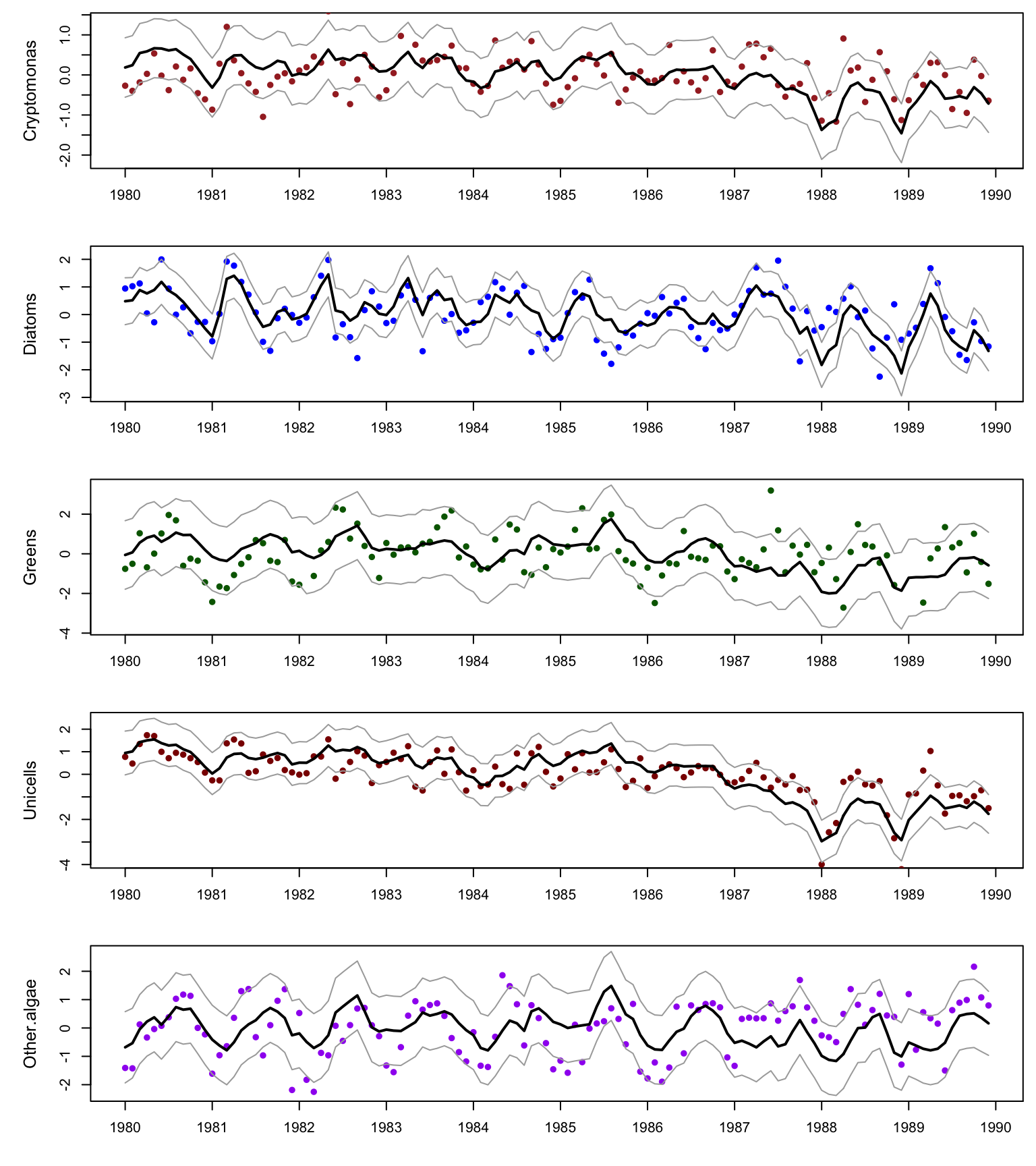
Figure 10.4: Data and fits from the DFA model.