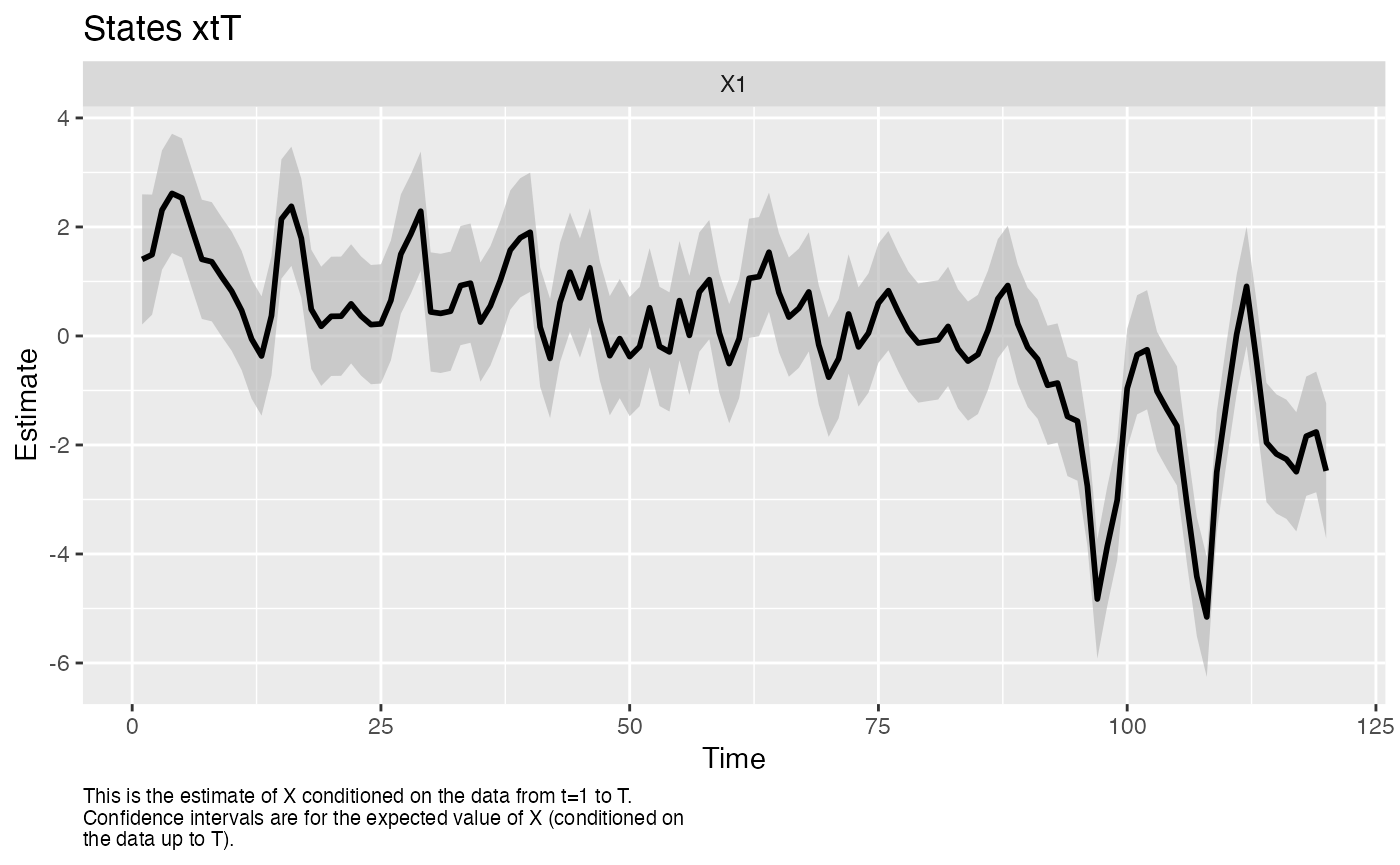
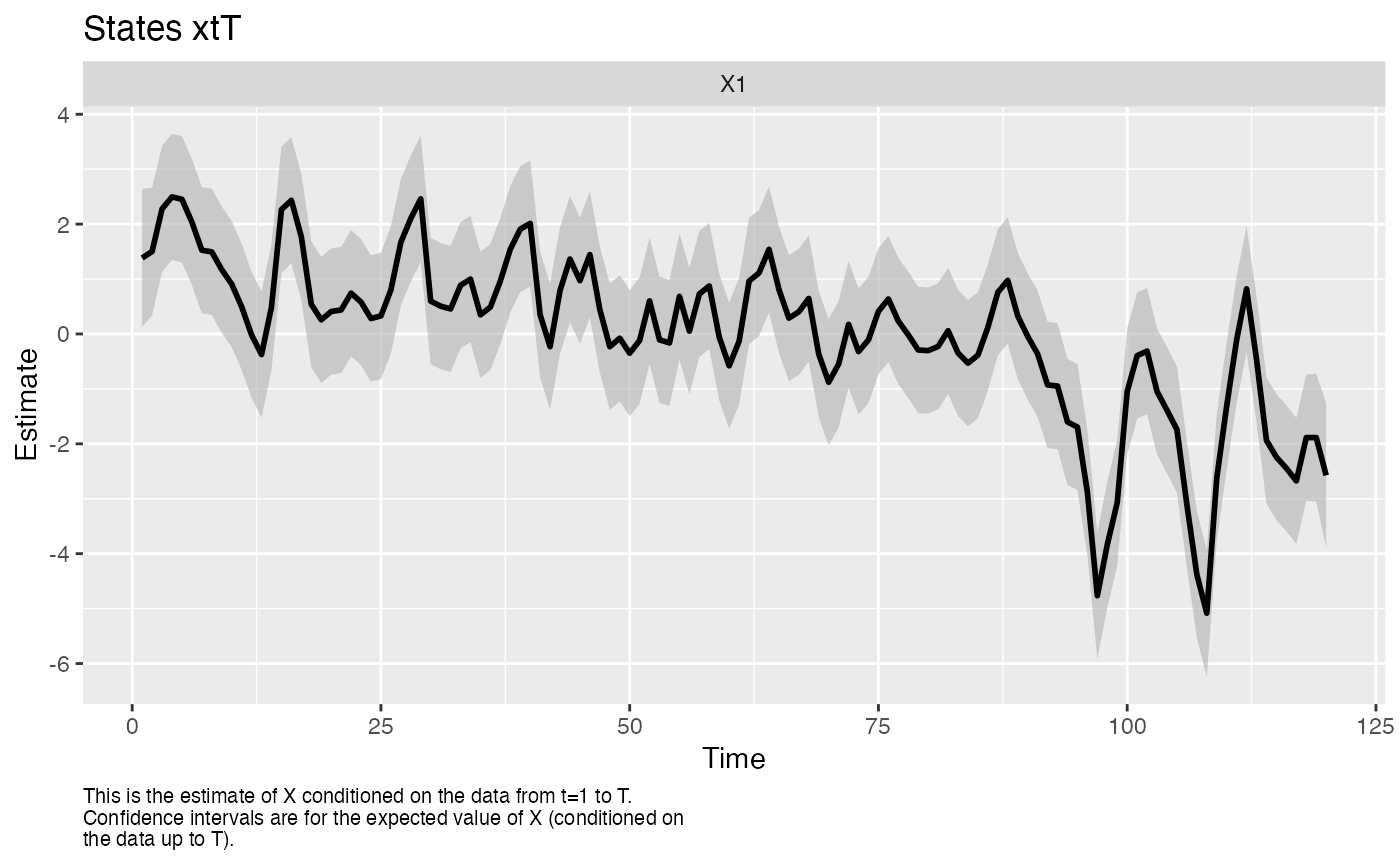
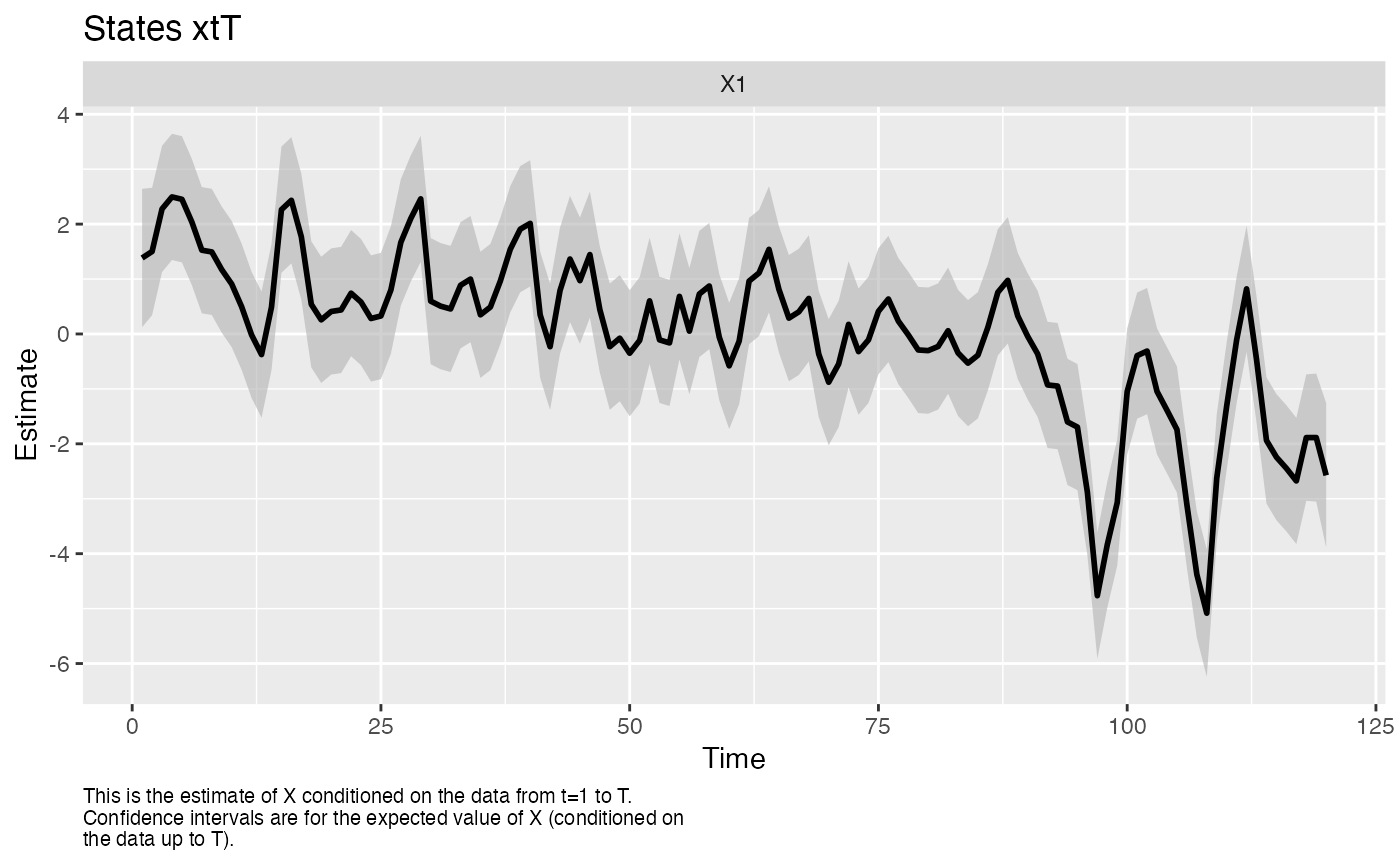
The MARSS() function allows you to fit DFAs with the
same form as MARSS(x, form="dfa"). This has a diagonal
\(\mathbf{Q}\) with 1 on the diagonal
and a stochastic \(\mathbf{x}_1\) with
mean 0 and variance of 5 (diagonal variance-covariance matrix). There
are only 3 options allowed for \(\mathbf{R}\): diagonal and equal, diagonal
and unequal, and unconstrained.
Example data
library(MARSS)
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
data(lakeWAplankton, package = "MARSS")
phytoplankton <- c("Cryptomonas", "Diatoms", "Greens", "Unicells", "Other.algae")
dat <- as.data.frame(lakeWAplanktonTrans) |>
subset(Year >= 1980 & Year <= 1989) |>
subset(select=phytoplankton) |>
t() |>
MARSS::zscore()Fit models without covariates
mod.list <- list(R='unconstrained', m=1, tinitx=1)Fit with MARSS with EM or optim and BFGS.
m1 <- MARSS(dat, model=mod.list, form='dfa', z.score=FALSE, silent = TRUE)
m2 <- MARSS(dat, model=mod.list, form='dfa', z.score=FALSE, silent = TRUE, method="BFGS")Fit with TMB.
library(marssTMB)
m3 <- dfaTMB(dat, model=list(m=1, R='unconstrained'))
m4 <- MARSS(dat, model=mod.list, form='dfa', method="TMB", silent=TRUE)
m5 <- MARSS(dat, model=mod.list, method="BFGS_TMB", form='dfa', silent=TRUE)Add example with covariates
For form="dfa", pass in covariates with
covariates=xyz. If using the default model form (not dfa),
then pass in covariates with model$d or
model$c.
Fit model
# use a simpler R
mod.list2 <- list(m=1, R='diagonal and unequal', tinitx=1)
# add a temperature covariate
temp <- as.data.frame(lakeWAplanktonTrans) |>
subset(Year >= 1980 & Year <= 1989) |>
subset(select=Temp)
covar <- t(temp) |> zscore()
m.cov1 <- MARSS(dat, model=mod.list2, form="dfa", covariates=covar, silent = TRUE, z.score = FALSE, method="TMB")Add a 2nd covariate
Parameter estimates
Model with one covariate.
tidy(m.cov1)
#> term estimate std.error conf.low conf.up
#> 1 Z.11 0.31418922 0.05433970 0.20768536 0.42069309
#> 2 Z.21 0.23028343 0.05024996 0.13179531 0.32877154
#> 3 Z.31 0.15133602 0.04841083 0.05645254 0.24621950
#> 4 Z.41 0.56773162 0.06970745 0.43110752 0.70435571
#> 5 Z.51 -0.05856336 0.04136120 -0.13962982 0.02250309
#> 6 R.(Cryptomonas,Cryptomonas) 0.75260003 0.10172886 0.55321512 0.95198493
#> 7 R.(Diatoms,Diatoms) 0.77087944 0.10199558 0.57097177 0.97078711
#> 8 R.(Greens,Greens) 0.71350994 0.09469766 0.52790594 0.89911394
#> 9 R.(Unicells,Unicells) 0.20849764 0.05388283 0.10288923 0.31410604
#> 10 R.(Other.algae,Other.algae) 0.68761333 0.08891885 0.51333560 0.86189107
#> 11 D.Cryptomonas 0.05877344 0.09805646 -0.13341369 0.25096058
#> 12 D.Diatoms -0.27858516 0.09131446 -0.45755821 -0.09961210
#> 13 D.Greens 0.51108061 0.08392952 0.34658177 0.67557945
#> 14 D.Unicells 0.13121433 0.10950305 -0.08340770 0.34583636
#> 15 D.Other.algae 0.53925536 0.07722438 0.38789836 0.69061236Model with two covariates.
tidy(m.cov2)
#> term estimate std.error conf.low conf.up
#> 1 Z.11 0.34060027 0.05594385 0.23095234 0.450248196
#> 2 Z.21 0.26339330 0.05139875 0.16265360 0.364132998
#> 3 Z.31 0.18775387 0.04887535 0.09195995 0.283547783
#> 4 Z.41 0.54020043 0.06833001 0.40627607 0.674124789
#> 5 Z.51 -0.04027862 0.04035477 -0.11937251 0.038815271
#> 6 R.(Cryptomonas,Cryptomonas) 0.71286331 0.09804098 0.52070651 0.905020101
#> 7 R.(Diatoms,Diatoms) 0.72982361 0.09775673 0.53822393 0.921423282
#> 8 R.(Greens,Greens) 0.68092519 0.09112594 0.50232163 0.859528737
#> 9 R.(Unicells,Unicells) 0.23937755 0.05464675 0.13227188 0.346483222
#> 10 R.(Other.algae,Other.algae) 0.67572369 0.08731346 0.50459246 0.846854917
#> 11 D.(Cryptomonas,Temp) -0.07369866 0.12487871 -0.31845644 0.171059118
#> 12 D.(Diatoms,Temp) -0.40186330 0.11652894 -0.63025583 -0.173470764
#> 13 D.(Greens,Temp) 0.39488271 0.10539729 0.18830781 0.601457608
#> 14 D.(Unicells,Temp) 0.13345292 0.13402433 -0.12922993 0.396135772
#> 15 D.(Other.algae,Temp) 0.45802179 0.09598266 0.26989923 0.646144351
#> 16 D.(Cryptomonas,TP) -0.22323976 0.12563874 -0.46948716 0.023007644
#> 17 D.(Diatoms,TP) -0.20994256 0.11623684 -0.43776258 0.017877454
#> 18 D.(Greens,TP) -0.21065665 0.10654015 -0.41947151 -0.001841801
#> 19 D.(Unicells,TP) 0.01447560 0.13811710 -0.25622893 0.285180141
#> 20 D.(Other.algae,TP) -0.13975971 0.09523756 -0.32642189 0.046902477